Leveraging algorithms for comparison
DM
In match configuration features, many comparison types are available to improve matching in different situations. For example, because of how it compares strings, the nGram comparison can find matches that Jaro-Winkler cannot. You should use both comparison types when matching names.
Review the available algorithms to understand which type to use for particular data.
The Bag of Words algorithm divides two strings of characters into a list of words and computes a score based on the similarity between the two strings. It is similar to the nGram comparison, except that it uses words, instead of grams as the segmentation. It ignores word order, but it is more sensitive to misspellings.
When you use the Bag of Words algorithm in Basic mode, you can define the following settings:
- Language Structure - Segments the strings for easier comparisons.
- Latin (All other languages) - Segments the strings based on white space and removes characters that are included in the Ignore Punctuation setting.
- CJK (Chinese, Japanese, or Korean characters) - Creates single character segments.
- Ignore Punctuation - Removes punctuation characters from the string.
- Threshold - Determines the confidence level for the comparison. If the algorithm has a threshold of 0.82, any string pairing with a score of 0.82 or higher is considered a match.
Bag of Words score
Bag of Words compares words in a string to determine how similar they are and computes a score as follows:
- 2 * (# of unique words appearing in both strings) / ((# unique words in first string) + (# unique words in second string))
The following sample string would be treated as follows:
Incoming record: Department of Cardiology Salem General Hospital
Veeva OpenData record: Salem General Department of Cardiology
- # of unique words appearing in both strings = 5
- # of unique words in the first string = 6
- # unique words in second string= 5
- Score = 2 * 5 / (6 + 5) = 0.90
This same comparison using different algorithms results in the following scores:
- Jaro-Winkler = 0.87
- n-Gram = 0.86
Equal comparison
Equal comparison checks if two values are exactly the same. Using this algorithm enables you to create flexible match features.
You can compare data four ways using equal comparison.
- FLOATS - Attempts to convert input strings to 8 byte IEEE 754 floating point numbers and compares them. If the conversion fails the comparison returns false.
- INTEGERS - Attempts to convert the strings to signed 64-bit integers and compares them. If the conversion fails, the comparison returns false.
- OBJECTS - Case-sensitive string comparison. The comparison fails if the strings contain different uppercase and lowercase letters.
- STRINGS - Compares strings without case-sensitivity.
The default comparison method is STRINGS.
| STRINGS | INTEGERS | FLOATS | OBJECTS | |
| "123" and "0123" | false | true | true* | false |
| "123.0" and "0123.0" | false | false | true | false |
| "Abc" and " abc " | true | false | false | false |
| "abc" and "abc" | true | false | false | true |
Note: Floating point math can round large numbers.
Example: Advanced mode syntax
When you define the comparison method, (for example, STRINGS) always use the following:
-
uppercase
-
plural
<equalComparison>
<compareAs>STRINGS</compareAs>
</equalComparison>
Jaro-Winkler
The Jaro-Winkler algorithm determines the similarity of two strings.
When you use the Jaro-Winkler algorithm in Basic mode, you can define the following settings:
- Include Winkler portion of algorithm - Using the Winkler portion increases the score if the first four characters are the same.
Winkler is useful for matching human names. It might not be appropriate for HCO names. For example, hospitals that are named after different saints will have the first four characters in common, so Saint James and Saint Paul will be considered very similar.
- Names should match - Enabling this increases the probability of a match when the number of matched characters is large. In Advanced Mode, this setting is
<usingLargeStringTolerance>. - Threshold - Fields with comparison scores above the threshold are considered equal; for example, if the algorithm has a threshold of 0.82, any string pairing with a score of 0.82 or higher is considered a match.
For examples of using the Jaro-Winkler algorithm in Basic and Advanced Mode, see Defining features.
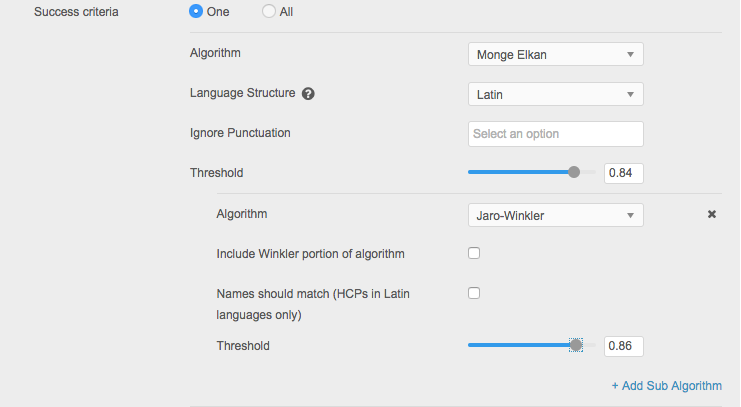
Monge Elkan comparison
The Monge Elkan algorithm is a hybrid string comparison. It first breaks the strings into tokens using the segmentation scheme, and then tries to find an optimal alignment between the tokens. The default token comparison method is to check if one of the tokens is a prefix of the other; for example hosp and hospital.
When you use the Monge Elkan algorithm in Basic mode, you can define the following settings:
- Language Structure - Segments the strings for easier comparisons.
- Latin (All other languages) - Segments the strings based on white space and removes characters that are included in the Ignore Punctuation setting.
- CJK (Chinese, Japanese, or Korean characters) - Creates single character segments.
- Ignore Punctuation - Removes punctuation characters from the string.
- Threshold - Determines the confidence level for the comparison. If the algorithm has a threshold of 0.82, any string pairing with a score of 0.82 or higher is considered a match.
- Add Sub Algorithm - Define another algorithm to use with Monge Elkan to improve the match outcome. All other algorithms are available to use.
nGram comparison
The nGram algorithm divides two strings of words into n-grams and computes a score based on the similarity between the two strings.
When you use the nGram algorithm, you can define the following settings:
- String length - Determines how many characters the string will be divided into (so a string length of 2 indicates that the string is broken into parts that are two characters in length each).
- Threshold - Determines the confidence level for the comparison. If the algorithm has a threshold of 0.82, any string pairing with a score of 0.82 or higher is considered a match.
The thresholds for nGram are considerably different than for Jaro-Winkler; a threshold of 0.50 is a good starting point, along with a Gram length of 2. Either setting can be modified to suit a particular incoming data set.
nGram score
nGram compares parts of a string to determine how similar they are and computes a score as follows:
- 2 * (# of unique n-grams appearing in both strings) / ((# unique n-grams in first string) + (# unique n-grams in second string))
Using nGram comparison, the following sample string would be treated as follows (spaces are removed prior to calculation):
Incoming record: Hospital Ryder Memorial
2-Grams =HO OS SP PI IT TA AL LR RY YD DE ER RM ME EM MO OR RI IA AL
Veeva OpenData record: Ryder Memorial Hospital
2-Grams = RY YD DE ER RM ME EM MO OR RI IA AL LH HO OS SP PI IT TA AL
- # of unique n-grams appearing in both strings = 19
- # of unique n-grams in the first string = 20
- # unique n-grams in second string= 20
- Score = 2 * 19 / (20 + 20) = 0.95
This same comparison using Jaro-Winkler results in a low score of 0.74.
The following example shows nGram comparison used in a feature:
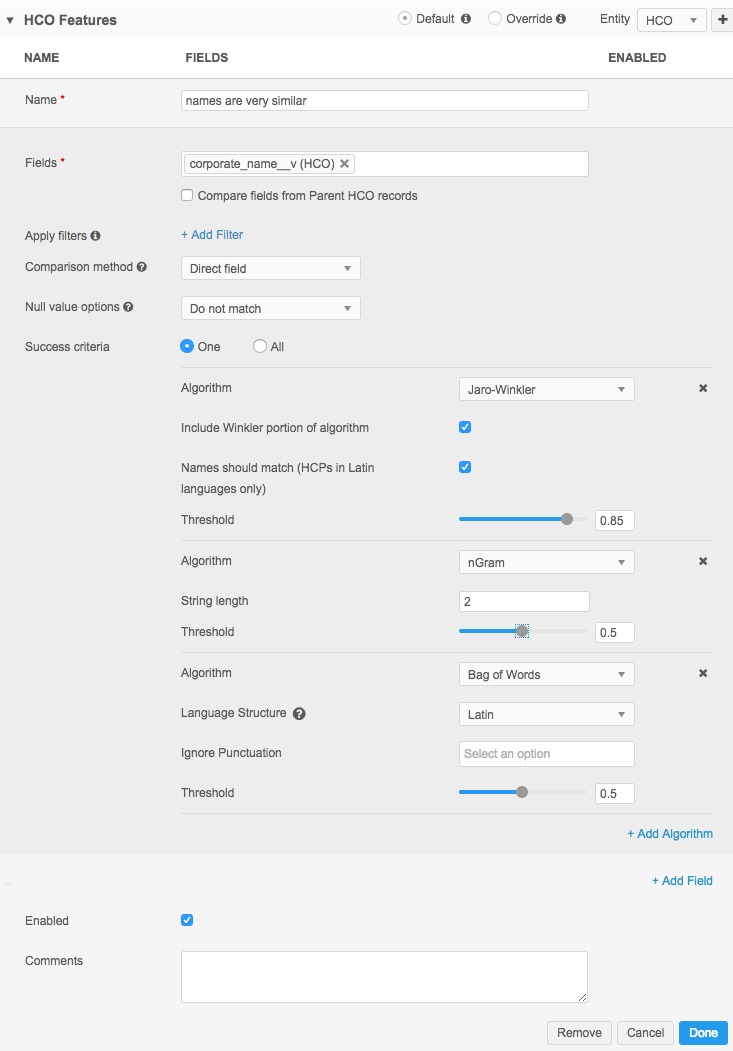
In Advanced Mode, the comparison looks like this:
<feature>
<name>names are very similar using nGram</name>
<enabled>true</enabled>
<comments></comments>
<collate>
<direct>
<field>corporate_name__v</field>
<nullMatching>IGNORE</nullMatching>
<nGramComparison>
<gramLength>2</gramLength>
<threshold>0.50</threshold>
</nGramComparison>
</direct>
</collate>
</feature>
Concatenating fields
The nGram algorithm is useful for concatenating name fields.
Network stores US name data in individual fields: first_name__v, middle_name__v and last_name__v, but different systems can store names in different fields. For example, maiden names are sometimes stored in the middle name field. Concatenating the name fields can assist in properly matching records where the names are broken up differently across the three name fields.
The following example illustrates how you can use concatenation in match rules. You should use nGram comparison to ensure accurate results. The confidence values needed for this configuration should be set much higher than if these fields were compared individually. You should analyze the outcomes of this rule carefully to ensure overmatching is not occurring.
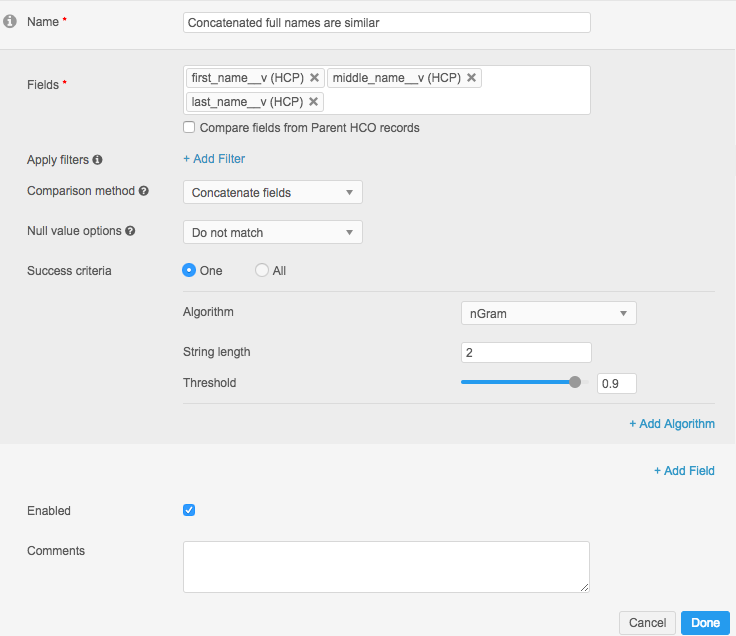
In Advanced Mode, the definition looks like this:
<feature>
<name>concatenated full names are similar</name>
<enabled>true</enabled>
<comments></comments>
<collate>
<concat>
<field>first_name__v</field>
<field>middle_name__v</field>
<field>last_name__v</field>
<nullMatching>STRICT</nullMatching>
<nGramComparison>
<gramLength>2</gramLength>
<threshold>0.90</threshold>
</nGramComparison>
</concat>
</collate>
</feature>
Phonetic comparisons
Match configuration features support several phonetic comparisons. Each comparison checks for equality of encoded strings. Choose the algorithm that works best with the data that you are matching.
The phonetic comparisons are:
- Beider Morse - Designed to compare Slavic and Yiddish names.
- Double Metaphone - Designed to compare English words or names that sound similar.
- NYSIIS (New York State Identification and Intelligence System) - Designed to match names by sound, as pronounced in English, despite minor differences in spelling.
Pinyin jaro
The Pinyin Jaro algorithm compares Chinese character strings using Hanyu Pinyin as a measure of character equivalence and the Jaro algorithm for overall string similarity.
Pinyin Jaro can be used in match configurations in both Basic and Advanced Mode.
Substrings
Use the Substring algorithm (substringComparison element in Advanced Mode) to check for matches between strings of two values. It can identify a substring starting within a string of text, or extract a substring from the end of a string of text. This option supports using an offset with a negative (-) number which starts the match at the end or middle of the string, depending on the length specified.
The following settings are used for the substring algorithm:
- Starting character - Specifies the character to begin with. The first character of a string is 0 (zero). The last character of a string is -1. In Advanced mode, this setting is called
<offset>. Open the Tooltip to see an example for defining the offset.
to see an example for defining the offset. - Length - Specifies the length of the substring to use for comparison.
The substring algorithm is particularly useful to use with other algorithms because it can make two records consistent in content so they can be compared to each other. middle_name__v field, while other countries use a full middle name. Use the substring comparison to extract the common string value (the middle initial) so that the two records can be now compared with the same content (first name, middle initial, last name).
The following example considers the middle name as an initial, so the substring length is one character (the initial) and it starts at the beginning of the string (Starting Character is 0).
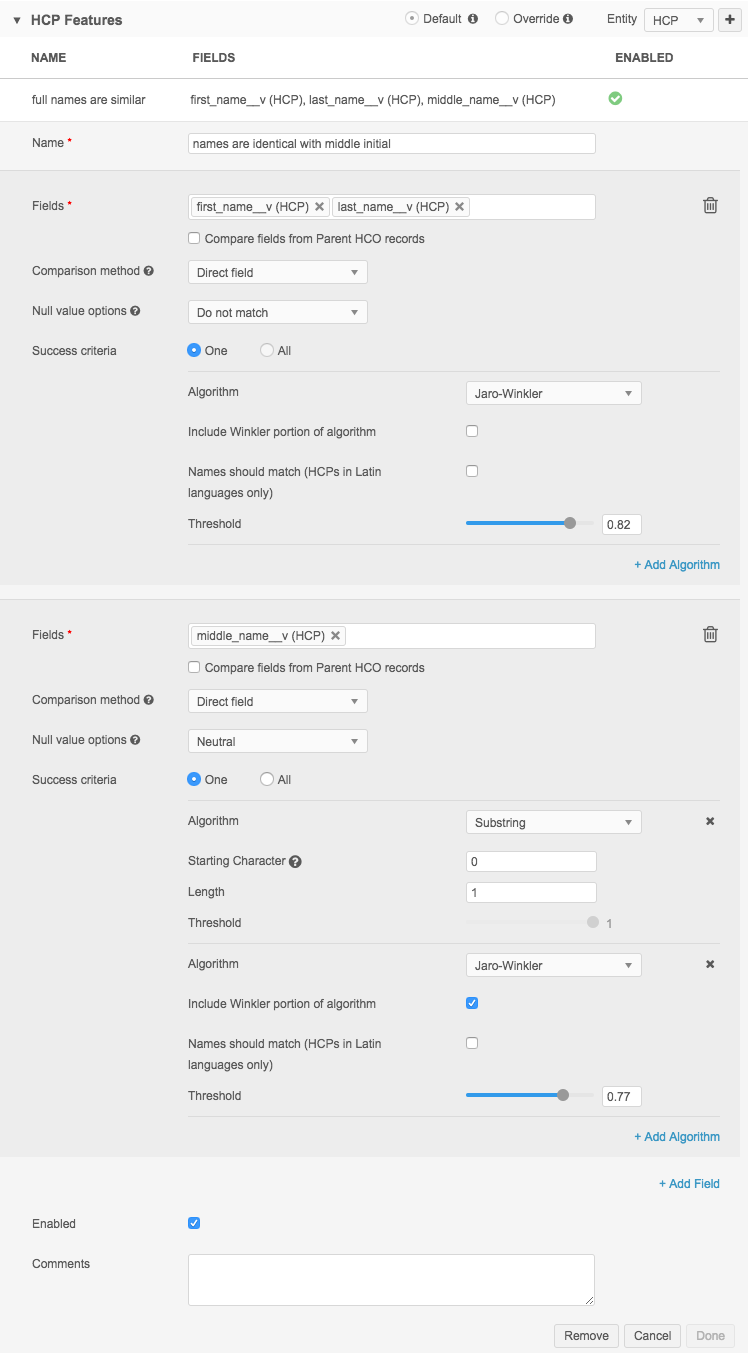
The definition looks like this in Advanced Mode:
<feature>
<name>names are identical with middle initial</name>
<enabled>true</enabled>
<comments></comments>
<collate>
<direct>
<field>first_name__v</field>
<field>last_name__v</field>
<nullMatching>STRICT</nullMatching>
<jaroWinklerComparison>
<usingWinklerExtention>false</usingWinklerExtention>
<usingLargeStringTolerance>false</usingLargeStringTolerance>
<threshold>0.83</threshold>
</jaroWinklerComparison>
</direct>
<direct>
<field>middle_name__v</field>
<nullMatching>IGNORE</nullMatching>
<multiComparison>
<agreement>ONE</agreement>
<substringComparison>
<offset>0</offset>
<length>1</length>
</substringComparison>
<jaroWinklerComparison>
<usingWinklerExtention>true</usingWinklerExtention>
<usingLargeStringTolerance>false</usingLargeStringTolerance>
<threshold>0.83</threshold>
</jaroWinklerComparison>
</multiComparison>
</direct>
</collate>
</feature>
Using negative substrings
When you use negative substrings, an offset of -1 starts at the last character of the string. An offset of -4 will start at the fourth last character of the string. The length considers the number of characters from that point backwards in the string.
<substringComparison> <offset>-1</offset> <length>4</length> </substringComparison>
Examples:
- XXXjohn and YYjohn would be equal using the substring comparison with offset = -1 and length = 4
- XXXjohnX and YYjohnY would be equal with offset = -2 and length = 4.
Matching on dates
Network supports matching on date fields. Using the substring matching algorithm with an offset of 0 (zero) and length of 10, dates can be matched using normal match rules. Dates must be formatted as yyyy-mm-dd, or they need to be transformed into that format during load.
Example
This is an example of a feature used for matching dates for the Grad Training Start Date (grad_trg_start_date__v) HCP field.
<feature>
<name>Dates are identical</name>
<enabled>true</enabled>
<comments></comments>
<collate>
<direct>
<field>grad_trg_start_date__v</field>
<nullMatching>IGNORE</nullMatching>
<substringComparison>
<offset>0</offset>
<length>10</length>
</substringComparison>
</direct>
</collate>
</feature>
Using multiple comparison methods
Features can use one or multiple algorithms. To use more than one algorithm at a time, you can add algorithms to the feature definition. Using this method, you can include any number of algorithms to match on, including one or all of them.
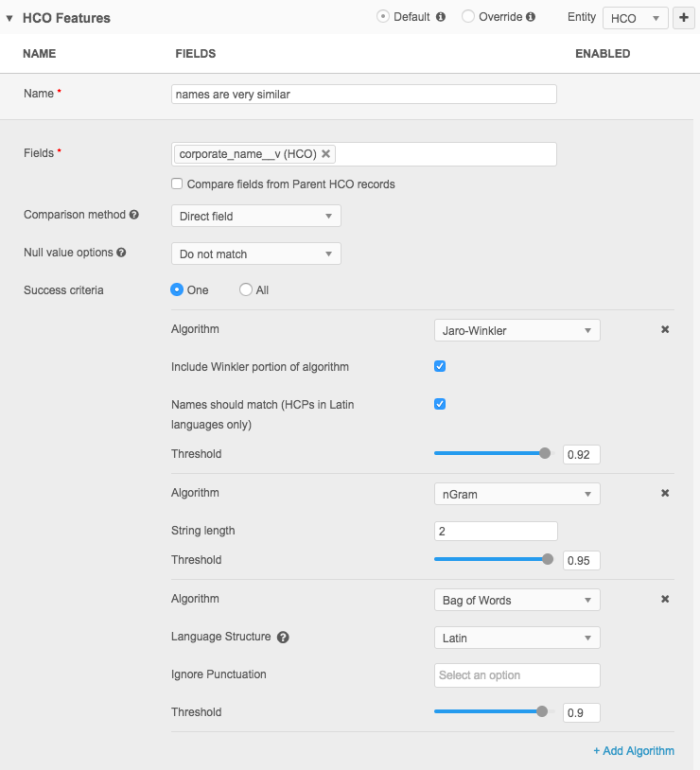
The default for the agreement option is ONE, but ALL can also be used. ONE means that as long as at least one of the algorithm passes, the feature will pass. ALL means that all specified algorithms must pass for the feature to pass.
In Advanced mode, you use the <multiComparison> element.
<feature>
<name>name is very similar</name>
<collate>
<direct>
<field>corporate_name__v</field>
<nullMatching>STRICT</nullMatching>
<multiComparison>
<agreement>ALL</agreement>
<jaroWinklerComparison>
<threshold>0.85</threshold>
</jaroWinklerComparison>
<nGramComparison>
<threshold>0.95</threshold>
<gramLength>2</gramLength>
</nGramComparison>
<bagOfWordsComparison>
<threshold>0.90</threshold>
</bagOfWordsComparison>
</multiComparison>
</direct>
</collate>
</feature>